Mycobacteria
Mycobacterium Testing By PCR
Sampling: swabbing.
Frogs were swabbed utilizing Medical Wire & Equipment Co (UK) MW 100-100 (Figure 1). Swabbed frogs were caught, isolated, identified and weighed before swab samples were taken aseptically from the posterior (back) and anterior (stomach) sides together with the undersides of feet and legs. The swabbing was done comprehensively repeating 2 or 3 times per area. The samples were then stored at 4°C to inhibit growth of other microorganisms. The largest problem associated with specimen collection is known to be contamination, thus care was taken to avoid cross contamination. This was done by changing gloves when handling different frogs, changing buckets and water regularly, and storing samples as soon as taken.
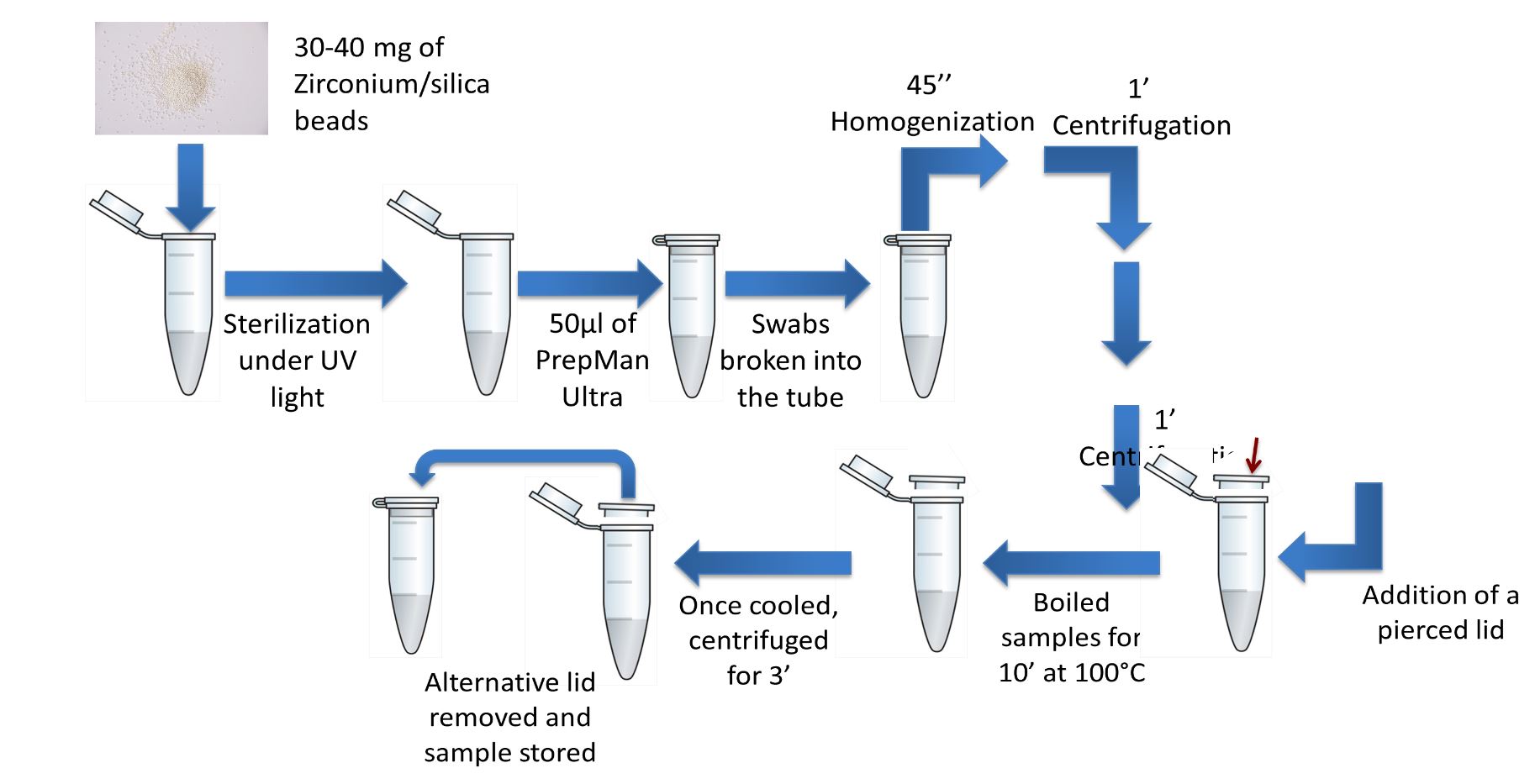
Extraction and homogenization of nucleic acids from samples.
30 to 40 mg of Zirconium/silica beads (Biospec. Products) measuring 0.5 mm diameter were weighed into separate Eppendorf centrifuge tubes. These were sterilized by placing them for 5 minutes under a UV light and 50ml of PrepMan Ultra (Applied Biosystems) was then added to each tube.
Swabs were broken into each labeled 1.5ml Eppendorf tube and homogenized for 45 seconds in a Mini Beadbeater 8, followed by 1-minute centrifugation at 13000 x gm in a microfuge. The homogenization and centrifugation was repeated to recover all material at the bottom of the tube.
The homogenized sample was placed in a heat block for 10 minutes at 100°C. An additional pierced lid was employed at this step (Figure 21). This is to trap the vapor formed while the sample boils but allows some steam off to release pressure. After 2-3 minutes of cooling to allow the condensation to liquidize, the sample was once again centrifuged at 13000 x gm for a further 3 minutes. Pierced lids were then removed.
The homogenized samples were stored in the fridge until further use.Figure 21. Diagrammatic representation of sample preparation, i.e. DNA homogenization. Note the red arrow highlights the use of a spare eppendorf lid previously perforated with a needle. The small hole is made as to decrease the pressure inside the eppendorf tube while sample is boiled.
The homogenized samples were stored in the fridge until further use.
Analysis of samples by PCR
Testing for mycobacterium with 95% probability of detecting 2% infected.
PCR reactions were prepared on individual tubes or 96 wells plates which contain 2 replicates of each sample, a positive control and a negative control. Wells were loaded with a total of 25ml. Specimen wells contained 24ml of Master mix (see table) and 1ml of DNA. The positive control had 24ml of Master mix and 1ml positive DNA. The negative controls were only 24ml Master mix and 1ml of dH2O.
The amplification conditions utilized were: 2 min 50°C, 10 min 90°C, 15s 95°C, 1 min 60°C for a total of 50 cycles.
PCR solutions
Table 1. Standard Master Mix used in PCR for 1 reaction
| Component | Volume | Total Volume |
| PCR Master mix | 12.5ml | 25ml |
| Primer (Forward) | 0.5ml | |
| Primer (Reverse) | 0.5ml | |
| dH2O (ultra pure – sigma) | 10.5ml | |
| DNA | 1ml |
Dilution of stock solutions
Table 2. Solutions and appropriate dilution uses in the Mycobacterium PCR Assay
| Solution | Stock concentration | Dilution | Final concentration |
| DNA sample | 1 | none | 1/1 |
| Primer (Forward) | 100mM | 25ml of Primer
475ml of dH2O |
5mM |
| Primer (Reverse) | 100mM | 25ml of Primer
475ml of dH2O |
5mM |
Primers Sequences:
Tb11
5’ ACC AAC GAT GGT GTG TCC AT 3’
Tb12
5’ CTT GTC GAA CCG CAT ACC CT 3’







