pTransgenesis
Introduction
With the Amaya Lab, we have developed a re-combinational cloning based on the Gateway® system, which we have named pTransgenesis, to overcome the limitations of traditional cloning methods. The Gateway® system utilizes in vitro site-specific recombination, which offers a rapid and convenient way of construction complex, multi-element vectors without the need for restriction enzymes and ligases. To this end, we adapted the Multi-site Gateway Three Fragment Vector Construction Kit® (Invitrogen, La Jolla, USA) which is commercially available and allows the assembly of three different constructs (entry clones) into a destination vector. We refer to the three entry vectors as position 1 (p1), containing a selection marker flanked by attL4-attR1 Gateway recombination sites; position 2 (p2), containing a promoter flanked by attL1-attL2 sites; and position 3 (p3), containing a reporter or coding sequence flanked by attR2-attL3. The destination vector, containing an I-SceI site and two Tol2 elements flanking the recombination cassettes, is referred to as position 4 (p4).
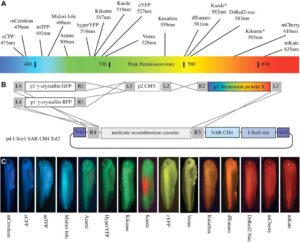
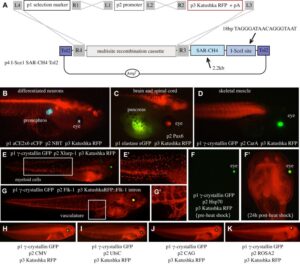
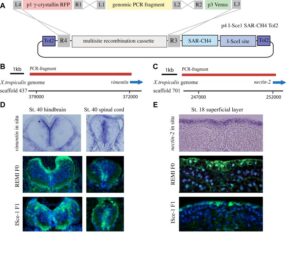
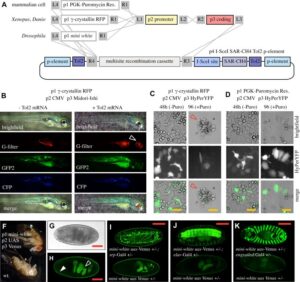
Design and construction of pTransgenesis vectors
To make a p1, p2, p3 entry clone, one can amplify a sequence of interest via PCR with primers, which contain sequences compatible with a subsequent “BP” recombination (another recombinase, BP Clonase II) with a p1, p2, p3 “DONR” vector (Invitrogen). Sometimes, however, it is easier is to use traditional subcloning methods to rearrange or redesign an existing p1, p2, p3 clone. In the case of the p2 position, a PCR product with TA overhangs can be cloned directly using a TOPO-based kit from Invitrogen (pCR®8/GW/TOPO® TA Cloning Kit). This kit renders promoter cloning and testing highly efficient as it allows recombination with selection and transgene coding sequences found in p1 and p3 vectors.
pTransgenesis LR recombination reactions
A critical point for the LR reaction is adding the correct molar amounts of the four vectors. We make “working stocks” of each vector such that 1µl of a p1, p2, and p3 working stock contains 10 fmols, and for p4 vectors, 20 fmoles. These dilutions must be performed in TE pH 8.0 (NB we also dissolve the originally prepped midi or maxi DNA pellets in TE pH 8.0). For the reaction, you’ll need Invitrogen’s “LR Clonase II Plus“ enzyme.
[/vc_column_text][vc_column_text]Working stocks can be calculated from the following formula (remember that the average molecular weight of 1-bp DNA is 660g/mol and hence 660fg/fmol):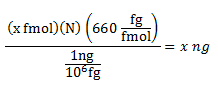
When:
N = bp length of a given plasmid
x = fmols required (10fmoles for p1,p2,p3: 20fmoles for p4)
For example, to make a p1 working stock of a 3333 bp plasmid:
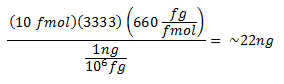
- Add the following to a 1.5-ml microcentrifuge tube: p1, p2, p3, p4 working stocks… 1 µl each = 4µl total TE pH 8.0, 4 µl
- Mix and then take 4µl of this 8µl mixture into a 1.5-ml microcentrifuge tube and add 1µl of LR Clonase II Plus enzyme, vortex, spin down, and incubate the reaction at ~23 degrees C overnight (NB: we store aliquots of the enzyme at -80°C).
- Add 0.5 μl of the Proteinase K solution to the reaction. Incubate for 10 minutes at 37˚C.
Transform a suitable E. colI host and spread on an ampicillin containing plate. We routinely use 2.75 µl of the 5.5 µl reaction for transformation into chemically competent OneShot TOP10 or DH5a cells (Invitrogen).
For plasmids ~5-15kb, pick 2-4 colonies for culture and digestion analysis (usually a ~75-100% success rate). For plasmids larger than 20kb, a greater number of colonies (4 to 8) should be picked (expect a ~25 – 75% success rate) and more care should be taken during culture (NOTE: avoid prolonged incubation after bacterial growth plateau).
Analysis of clones
Using DNA software one can easily predict the resulting recombinant DNA. We use a shareware called ApE (a plasmid editor,http://biologylabs.utah.edu/jorgensen/wayned/ape/). Using this elegant program, p1, p2, p3 and p4 clones can be recombined using the “Recombination Tool” from Tool tab. Chose the recombination type as Multi-site LR reaction, and select your entry clones in order. By referring to the sequence of the recombinant, one can analyse the LR generated clones using appropriate restriction enzymes. For large plasmids, we generally use BamHI or XbaI for digestion analysis.
 Note: This information comes from the Invitrogen manual found here.
Note: This information comes from the Invitrogen manual found here.


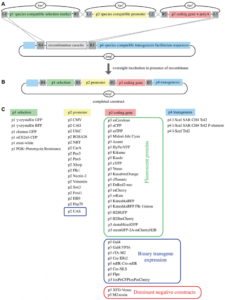
 A list of the whole collection can be viewed
A list of the whole collection can be viewed 




